|
Arrays with a block cyclic distribution were mentioned earlier, and in particular with a 1D block cyclic distribution, a vector (or block vector) may be split into blocks of equal size, given by a blocking factor and the blocks are then asigned to a vector of processes in cyclic manner. The concept is simple to extend to an N-dimensional array distributed over a process array of the same dimensionality.

|
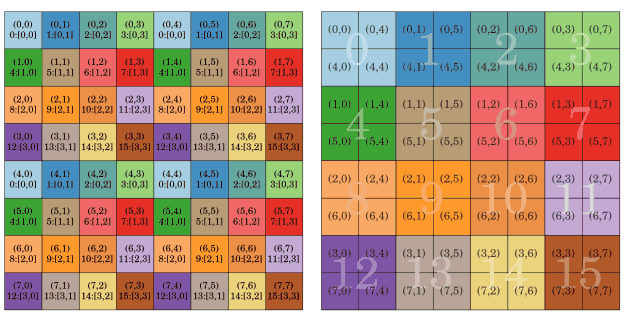
ScaLAPACK works with such 2D arrays and the spread of a square matrix on a square process array is shown in Fig 5. Operations are performed on the blocks with a single process, single or multithreaded, routines from BLAS and LAPACK. The distributed storage format reduces both communication and data duplication.
A generic wrapper routine was written which mimics closely the 'advanced' type diagonalisation drivers from ScaLAPACK with a few practical deviations. The routine manages all work arrays necessary internally and handles all combinations of { ,p}{dsy,zhe}{e,g}v{z,d,r} covering all cases of interest. It is also an interface to the diagonalisation routines in ELPA[7] since these use the same 2D block cyclic distribution as ScaLAPACK.
|
|
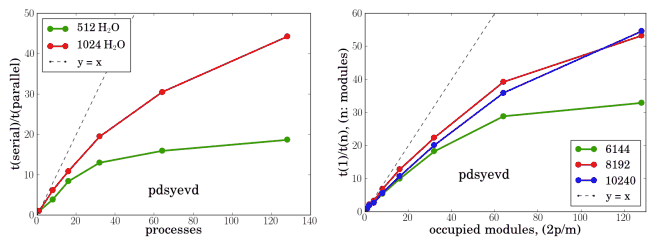
The ELPA library attempts to speedup the tridiagonalisation step (the bottleneck in diagonalisation) by replacing the BLAS2 *mv routines with BLAS3 *mm 6 at the cost of slower backsubstitution and the hope that not all eigenvectors will be required, which is the case in TB, especially for insulators. The optimal blocking factor was found to be 32.
The wrapper routine accepts 2D block cyclic distributed local arrays with respective descriptors and in the serial case calls the appropriate LAPACK routine instead of the parallel ones. This allows one to use exact wrappers or null libraries and thus produce single object file which can be packaged into a purely serial or parallel executable.
Since the Bloch transform is fast, local only and relatively simple even when one atom was spread across 2 or 4 processes and since the cubic scaling cost of diagonalisation with respect to the matrix dimensions a decision was made to not apply any padding and in this way minimise the size. The time for diagonalising genuine sparse Hamiltonian atoms or a randomly generated one is the same and any diagonal padding entries will increase the effort unnecessarily.
|